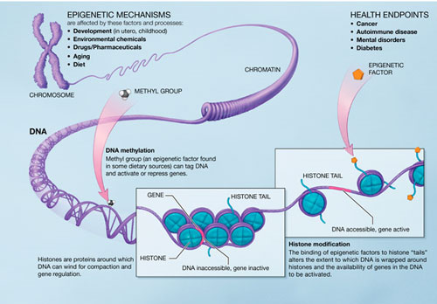
This image summarizes the main mechanisms of epigenetics. [Citation 18]
Two main mechanisms for epigenetic change exist: (1) DNA methylation and (2) and posttranslational histone modification (Citation 6).
DNA Methylation DNA methylation is the process by which a methyl group is added to the C5 position of the cytosine at a CpG site. CpG sites are spots in the genome in which a guanine follows a cytosine. The addition of the methyl group transforms the cytosine into 5' methylcytosine. Methylation follows general patterns which dictate which parts of the genome are methylated and which are not. For example, repetitive, non-coding sequences are heavily methylated. On the other hand, CpG islands (sequences with a high frequency of CpG sites) remain generally unmethylated, allowing the genes to be expressed (Citation 6). Molecular Process Methylation has the power to suppress genes, as the presence of a methyl group prevents transcription factors from functioning, thereby blocking transcription as a whole. The enzyme DNA methyltransferase adds the methyl group. After this has occurred, various proteins arrive and compact the DNA into heterochromatin. These molecules include methyl CpG binding protein 2 (MeCP2), histone deacetylases (HDAC), and histone methyltransferase (HMT). Such molecules serve to compact the euchromatin, a loosely wound form of chromatin, into heterochromatin, a tightly wound form of chromatin. The tightly wound nature of heterochromatin prevents the transcription factors from binding, thereby preventing DNA from copying and suppressing the genes in that region. (Citation 6) | 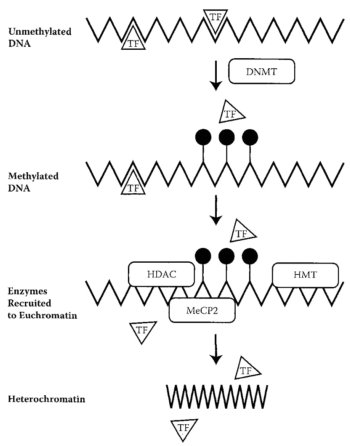 This diagram depicts the methylation of DNA and the consequences of this process. Refer to the following legend. TF-Transcription Factors DNMT-DNA Methyltransferase Filled Lollipops-Methyl groups MeCP2-Methyl CpG binding protein 2 HDAC-histone deacetylase HMT-histone methyltransferase (Citation 6)
|
Histone Modification Modifications to histones involve a complex aggregation of processes and reagents. This section will describe the foundations of modification, its general patterns, and the specific variants of modification. In the previous section, DNA methylation and its ability to alter the DNA, thereby inhibiting transcription was discussed. But how do histone modifications alter the genome? Histones are proteins which form the core of nucleosomes, the smallest unit of chromatin structure (Citation 28) . The manner in which the DNA in the nucleosomes interacts with the histones determines the overall state of the chromatin. Therefore, if the chromatin is not in its correct form, certain genes may be forced into an atypical state. The resulting phenotypic alteration is the result of the second major mechanism of epigenetic change: histone modification (Citation 6). The term "histone code" refers to the collective modifications to a histone". Before reading on, one must understand that histones are naturally modified. The phenotypic changes arise from modifications that have occured outside of the histone code (Citation 20). The Forms of Histone Modification Modifications to histones can alter the expression of genes in two ways: 1. Histones and DNA interact differently, causing the chromatin to coil incorrectly. 2. The altered histones signal for the presence of nonhistone proteins which alter the chromatin. First, the modification of a histone has the potential to alter the polypeptide's charge, which subsequently can alter the manner in which the histone and the nucleosome interact. The most pronounce and well-studied modification in this category is acetylation (look below for more information). The addition of an acetyl group to a lysine neutralizes the amino acid, thereby altering the overall structure of the chromatin. Typically, these variants of modification result in the unraveling of chromatin, which has gene expression implications. Phosphorylation, too, is known to alter histonal charges and chromatin structure, but this phenomenon has not been well studied. Generally speaking, though, "any alteration in histone charge will undoubtedly have structural consequences for the chromatin architecture" (Citation 24). Second, modifications to histones can signal for the binding of other proteins. This is accomplished through a protein domain's recognition of a specific modification. Domains are the base unit of tertiary protein structure. They are independent polypeptide chains that have the capability to fold into a stable protein. A domain's conformation recognizes a histone modification and binds to it. Categories of these proteins have been attributed to certain modifications. Acetylation, for example, is recognized by the bromodomains (Citation 24). Conversely, certain modifications can deter the binding of proteins. These exclusive capabilities may have implications in certain processes, notably transcription. Specific Modifications to Histones Histones may be modified through various chemical processes. The manner in which these these processes is executed, in addition to their effect, depends on the biological function under consideration. So, the role of acetylation in transcription is different from acetylation's role in DNA replication. The various modifications are discussed in this section. It is important to note, though, that epigenetics is a new field and that researches continue to discover new applications and areas of involvement for these modifications. For the purposes of clarity and comprehension, the following information only deals with some of the most well-known applications of specific histone modifications (Citation 24). Acetylation- This process, the addition of an acetyl group (COCH3) (Citation 3), is an important factor in gene activation or repression. Enzymes known as histone acetyltransferases (HATs) catalyze the addition of an acetyl group. Histone deacetyltransferases (HDACs) reverse that process. Acetylation correlation with activation of genes, while deacetylation tends to be related to their repression. Acetylation has also been connected to DNA replication. A HAT, HB01, appears to be crucial in a cell's progression into the S phase (the phase of division in which the DNA replicates) (Citation 24). Phosphorylation-This modification, which acts on serines and threonines, adds a phosphate (PO4) through the action of enzymes whose names end in -kinase (Citation 8). Conversely, a category of enzymes known as phosphatases reverse the process (Citation 29). This modification plays a significant role in DNA repair. Generally speaking, certain alterations to histones act as markers for regions of DNA needing repair. Phosphorylation plays this role in numerous histones (Citation 24). This modification variant also affects chromosome condensation, as the addition of a phosphate to specific sites dictates how the chromatin will compact. Ubiquitylation-This modification, which acts on the large scale, is the addition of a protein known as ubiquitin (Citation 21). Ubiquitilases add these polypeptide chains to lysines. Little is known about this modification, but research suggests that it recruits molecules to chromatin which preserve it in an open form. In addition, the ubiquitin, due to its relatively large size may itself maintain the chromatin in this form. In one case, enzymes have been discovered which prevent the ubiquitylation of a certain lysine on the H2 histone. The specificity of this mechanism, in addition to the lack of similar structures, limits the use of this modification. However, for this discovered instance, the deubiquitylation suggests that ubiquitin may be required for proper transcription (Citation 24). Environmental Influences Diet As seen in this video, diet can play a hugely important role in epigenetic changes. The mice that were fed a high methyl diet stayed healthy, while the mice that were fed a low methyl diet became extremely obese. Research has shown that different vitamins and nutrients can affect epigenetic mechanisms (Citation 4). One of the most promising finds, as demonstrated in the link above, is the relation between the nutrients folate (a B vitamin), vitamin B12, methionine and choline, and DNA methylation. These nutrients are all essential for humans, and are naturally found in our foods. B12 generally found in meats (generally in organs like the liver or kidney). Folate is also found in some cereals and leafy greens, while methionine, in addition to meats, is ubiquitous in cheeses and nuts. While these nutrients carry out numerous, essential functions, they play an important role in epigenetics-specifically in methylation. All four of these nutrients work to maintain and protect DNA structure, "preventing harmful alterations in the genetic code" (Fenech 119). By donating a carbon atom from their methyl groups, these nutrients effectively keep DNA "healthy" by synthesizing nitrogenous bases and by keeping cytosine unmethylated (Citation 4). Because all these nutrients act together, deficiency in one of them can lead to a breakdown of the methylation cycle. As the methylation cycle stops turning, uracil does not synthesize into uracil and methionine is unable to donate methyl groups to the DNMTs, resulting in less protection of the DNA. This in turn can lead to highly increased risks of diseases like cancer, obesity or diabetes. Consuming the suggested amounts of these everyday nutrients can effectively reduce epigenetic changes, while eating low methyl diets can significantly increase chances of genetic diseases (Citation 4). Other Factors Diet, however, is only one of the factors that can influence epigenetic changes. Research has shown that exposure to chemicals and even certain pharmaceuticals can interfere with the methylation cycle and can cause unwanted histone and miRNA modifications. Metals like nickel and cadmium can cause oxidative stress (an inability for the body to rid itself of hazardous intermediates) on DNA, which can "interfere with the ability of methyltransferases to interact with DNA thus resulting in a generalized altered methylation of cytosine residues at CpG sites." (Citation 2). Studies on the effects of chemicals is continuing to reveal more information about the potential epigenetic consequences. Another potential factor could be the lifestyle decisions of our parents during, and even before, our development. Olivier Rando, of the University of Massachusetts School of Medicine, conducted a study on the effects of fathers' diets. Using mice, Rando showed that low-protein diets in fathers, and even grandfathers, changed "hundreds of genes... in the offspring of those protein-starved males" (Citation 9). One interesting change that Rando noted was a histone modification, resulting in expression of a gene that controls lipid transcription factors in the liver. The children of the low-protein males lacked some of these factors, inhibiting their ability to break down lipids. The implications of the effects of lifestyle on epigenetics are huge. It could lead to the discovery of the causes of numerous diseases and disorders. | 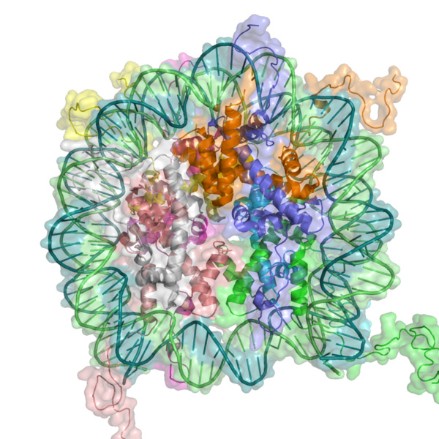 This computer-generated image portrays the wrapping of DNA around histones [Citation 27]. 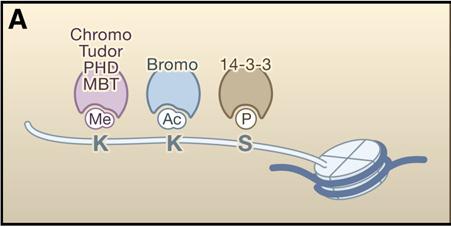 This diagram depicts the binding of protein domains to certain histone modifications, making clear the specificity of these interactions. Refer to the following legend. K-Lysine, S-Serine, Me-Methyl, Ac-Acetyl, P-Phosphate Methylation-This variant of methylation regards proteins, and therefore affects amino acids. Two variants of histone methylation exist: (1) lysine methylation and (2) arginine methylation. Both are accomplished by histone methyltransferases (HMTs), but each has their own specific variants. These enzymes, which add a methyl group, ordinarily modify a single amino acid on a single histone. Lysine methyltransferases catalyze lysine methylation. The addition of a methyl group (CH3) can have various effects. For example, depending on the site of addition, the methyl group can either activate or repress gene expression. In 2004, the first histone demethylase was discovered. These molecules have not been well investigated as of yet. Similarly to lysine methylation, arginine methylation may either activate or repress gene expression. Carried out by protein arginine methyltransferases (PRMTs), this process is generally cued by transcription factors. No enzymes which reverse this process have been identified and no proteins with domains that bind specifically to methylated arginines are known (Citation 24). Deimination-An imine is a a chemical compound which has a carbon double bonded to a nitrogen. They are present in certain amino acids. In the case of histone modifications, deimination occurs in the conversion of arginine to citrulline. This relates to gene expression, as citrulline can prevent arginine methylation. Therefore, if a certain arginine residue, which is normally methylated, becomes demethylated, then the overall form of the chromatin may be altered. This could potentially alter the transcription of the DNA. As of yet, no mechanism for the conversion of citrulline to arginine is known (Citation 24). 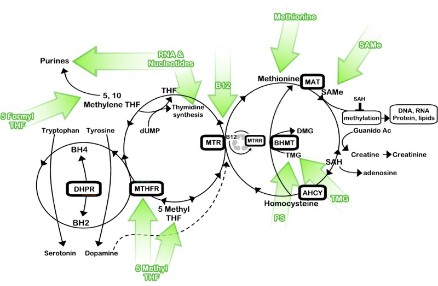 This picture depicts the methylation cycle and the importance of the four nutrients methionine, choline, folate and vitamin B12. We can see that folic acid (from diet, in the form of 5 formyl THF), is chemically broken down to power purine and pyrimidine synthesis. The folate also plays a role, along with vitamin B, in MS, or methionine synthesis. Methionine then becomes S-adenosyl methionine (SAM), which donates a methyl group to the DNMTs (DNA methyltransferases), which in turn methylate DNA. What is not depicted in this chart is choline, which, like B12, helps "turn" the MS wheel. (Citation 32) 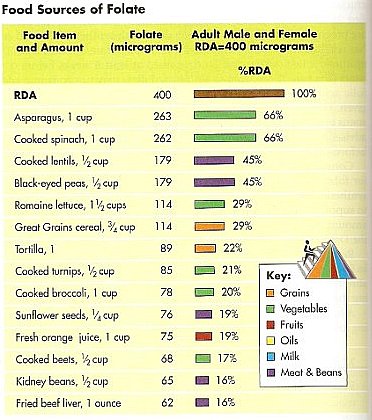 The above table details the quantities of folate contained within numerous foods, while the below table details the quantities of vitamin B12. Note: "RDA" stands for "Recommended Dietary Allowance". (Citation 34) 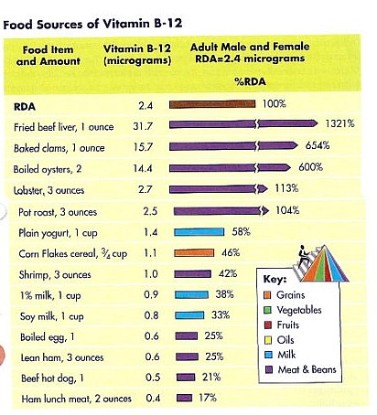 |